The environmental DNA qPCR hot start mix offers outstanding performance for sensitive, robust and precise probe-based qPCR detection and quantitation of target eDNA extracted from environmental samples. eDNA qPCR hot start mix is highly resistant to inhibiting factors, like humic acids in environmental samples.
The eDNA qPCR hot start mix has:
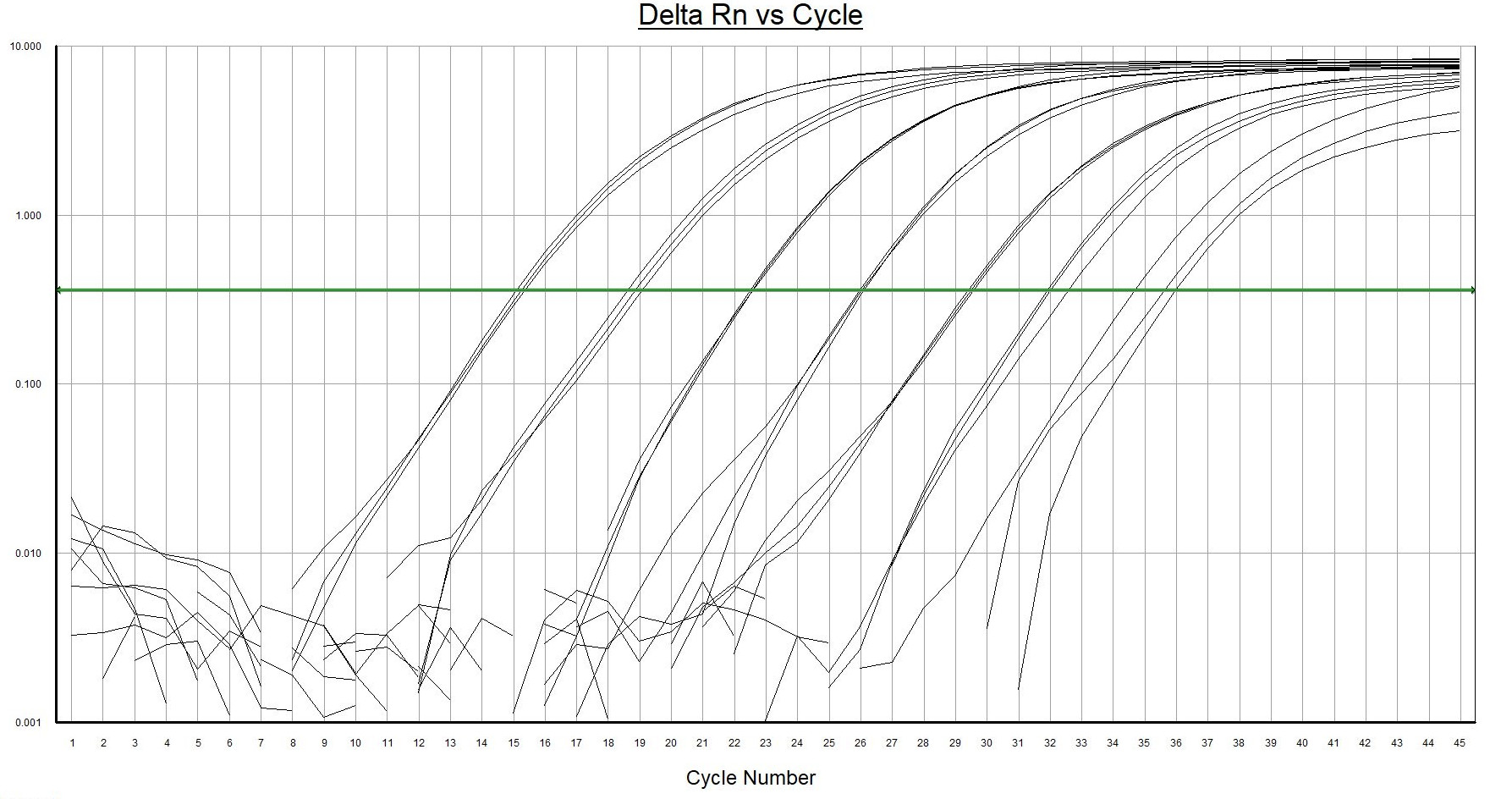
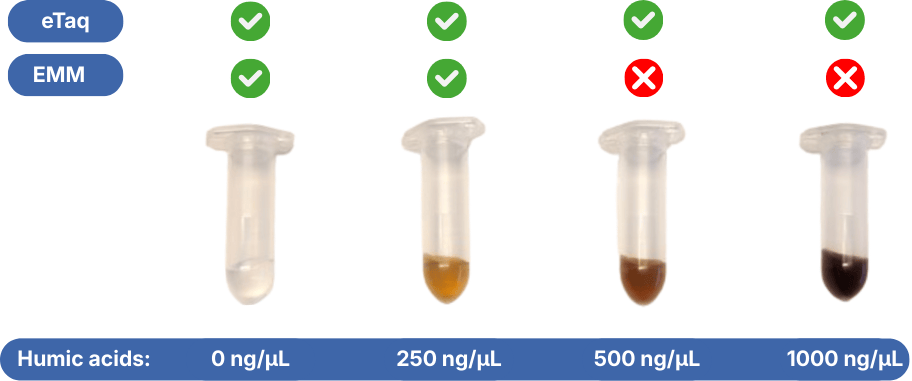
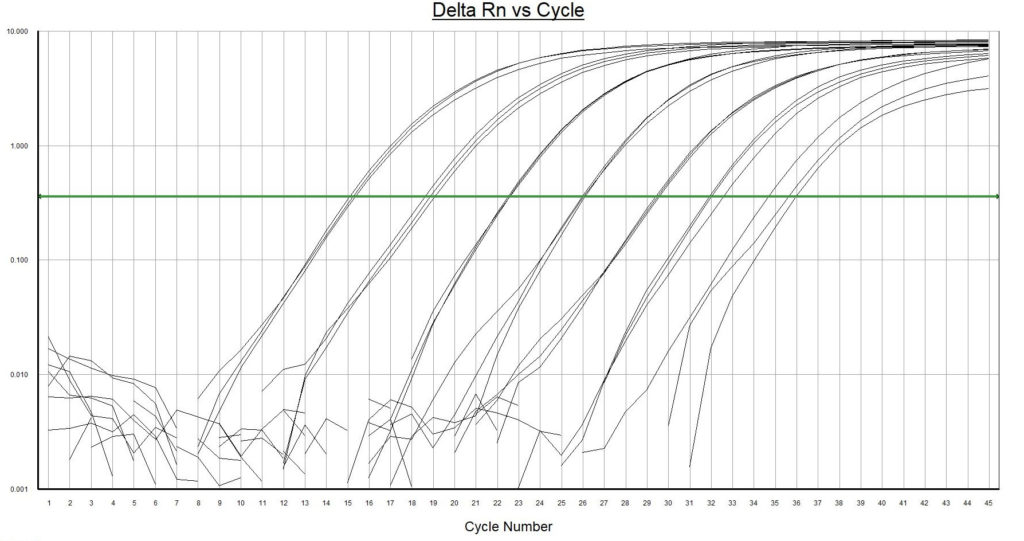
- Highest resistance to inhibiting factors in comparison to other qPCR master mixes. Environmental DNA extracts contain often multiple qPCR inhibiting factors. Normal qPCR master mixes are sensitive to these substances.
- Highest fluorescence signal with low background noise. Isolated environmental samples contain residues of naturally occurring auto fluoresce substances that will interfere with the measurements. A strong fluorescence signal from the analyses is required for this kind of samples.
- Highest possible sensitivity (1 DNA copy per reaction). Environmental water samples may contain very low amounts of target DNA.
- Hot-start properties. The DNA polymerase enzyme has been genetically modified to have similar or better hot-start properties when compared to antibody-mediated hot-start enzymes. Working on ice is not necessary during the preparation of the PCR mixture.
Kit content
- 2x 2500 µl eTaq qPCR mix (2x)
- 2x 200 µl eTaq DNA polymerase
Relevant Documents
- Protocol eDNA qPCR hot start mix (SYL1003)
- Validation report eDNA qPCR hot start mix (SYL1003)
- Material safety data sheets: Sylphium eTaq qPCR mix, Sylphium eTaq DNA polymerase
References
* Uchii, K., Doi, H., Okahashi, T., Katano, I., Yamanaka, H., Sakata, M. K., et al. (2019). Comparison of inhibition resistance among PCR reagents for detection and quantification of environmental DNA. DNA 1, 359–367.